The Case for Numba in Community Code
The numeric Python community should consider adopting Numba more widely within community code.
Numba is strong in performance and usability, but historically weak in ease of installation and community trust. This blogpost discusses these these strengths and weaknesses from the perspective of a OSS library maintainer. It uses other more technical blogposts written on the topic as references. It is biased in favor of wider adoption given recent changes to the project.
Let’s start with a wildly unprophetic quote from Jake Vanderplas in 2013:
I’m becoming more and more convinced that Numba is the future of fast scientific computing in Python.
– Jake Vanderplas, 2013-06-15
http://jakevdp.github.io/blog/2013/06/15/numba-vs-cython-take-2/
We’ll use the following blogposts by other community members throughout this post. They’re all good reads and are more technical, showing code examples, performance numbers, etc..
- https://flothesof.github.io/optimizing-python-code-numpy-cython-pythran-numba.html
- https://dionhaefner.github.io/2016/11/suck-less-scientific-python-part-2-efficient-number-crunching/
- http://jakevdp.github.io/blog/2013/06/15/numba-vs-cython-take-2/
- http://jakevdp.github.io/blog/2015/02/24/optimizing-python-with-numpy-and-numba/
- http://stephanhoyer.com/2015/04/09/numba-vs-cython-how-to-choose/
- https://murillogroupmsu.com/numba-versus-c/
At the end of the blogpost these authors will also share some thoughts on Numba today, looking back with some hindsight.
Disclaimer: I work alongside many of the Numba developers within the same company and am partially funded through the same granting institution.
Compiled code in Python
Many open source numeric Python libraries need to write efficient low-level code that works well on Numpy arrays, but is more complex than the Numpy library itself can express. Typically they use one of the following options:
- C-extensions: mostly older projects like NumPy and Scipy
- Cython: probably the current standard for mainline projects, like scikit-learn, pandas, scikit-image, geopandas, and so on
- Standalone C/C++ codebases with Python wrappers: for newer projects that target inter-language operation, like XTensor and Arrow
Each of these choices has tradeoffs in performance, packaging, attracting new developers and so on. Ideally we want a solution that is …
- Fast: about as fast as C/Fortran
- Easy: Is accessible to a broad base of developers and maintainers
- Builds easily: Introduces few complications in building and packaging
- Installs easily: Introduces few install and runtime dependencies
- Trustworthy: Is well trusted within the community, both in terms of governance and long term maintenance
The two main approaches today, Cython and C/C++, both do well on most of these objectives. However neither is perfect. Some issues that arise include the following:
- Cython
- Often requires effort to make fast
- Is often only used by core developers. Requires expertise to use well.
- Introduces mild packaging pain, though this pain is solved frequently enough that experienced community members are used to dealing with it
- Standalone C/C++
- Sometimes introduces complex build and packaging concerns
- Is often only used by core developers. These projects have difficulty attracting the Python community’s standard developer pool (though they do attract developers from other communities).
There are some other options out there like Numba and Pythran that, while they provide excellent performance and usability benefits, are rarely used. Let’s look into Numba’s benefits and drawbacks more closely.
Numba Benefits
Numba is generally well regarded from a technical perspective (it’s fast, easy to use, well maintained, etc.) but has historically not been trusted due to packaging and community concerns.
In any test of either performance or usability Numba almost always wins (or ties for the win). It does all of the compiler optimization tricks you expect. It supports both for-loopy code as well as Numpy-style slicing and bulk operation code. It requires almost no additional information from the user (assuming that you’re ok with JIT behavior) and so is very approachable, and very easy for novices to use well.
This means that we get phrases like the following:
- https://dionhaefner.github.io/2016/11/suck-less-scientific-python-part-2-efficient-number-crunching/
- “This is rightaway faster than NumPy.”
- “In fact, we can infer from this that numba managed to generate pure C code from our function and that it did it already previously.”
- “Numba delivered the best performance on this problem, while still being easy to use.”
- https://dionhaefner.github.io/2016/11/suck-less-scientific-python-part-2-efficient-number-crunching/
- “Using numba is very simple; just apply the jit decorator to the function you want to get compiled. In this case, the function code is exactly the same as before”
- “Wow! A speedup by a factor of about 400, just by applying a decorator to the function. “
- http://jakevdp.github.io/blog/2015/02/24/optimizing-python-with-numpy-and-numba/
- “Much better! We’re now within about a factor of 3 of the Fortran speed, and we’re still writing pure Python!”
- “I should emphasize here that I have years of experience with Cython, and in this function I’ve used every Cython optimization there is … By comparison, the Numba version is a simple, unadorned wrapper around plainly-written Python code.”
- http://jakevdp.github.io/blog/2013/06/15/numba-vs-cython-take-2/
- Numba is extremely simple to use. We just wrap our python function with autojit (JIT stands for “just in time” compilation) to automatically create an efficient, compiled version of the function
- Adding this simple expression speeds up our execution by over a factor of over 1400! For those keeping track, this is about 50% faster than the version of Numba that I tested last August on the same machine.
- The Cython version, despite all the optimization, is a few percent slower than the result of the simple Numba decorator!
- http://stephanhoyer.com/2015/04/09/numba-vs-cython-how-to-choose/
- “Using Numba is usually about as simple as adding a decorator to your functions”
- “Numba is usually easier to write for the simple cases where it works”
- https://murillogroupmsu.com/numba-versus-c/
- “Numba allows for speedups comparable to most compiled languages with almost no effort”
- “We find that Numba is more than 100 times as fast as basic Python for this application. In fact, using a straight conversion of the basic Python code to C++ is slower than Numba.”
In all cases where authors compared Numba to Cython for numeric code (Cython is probably the standard for these cases) Numba always performs as-well-or-better and is always much simpler to write.
Here is a code example from Jake’s second blogpost:
Example: Code Complexity
# From http://jakevdp.github.io/blog/2015/02/24/optimizing-python-with-numpy-and-numba/
# Numba # Cython
import numpy as np import numpy as np
import numba cimport cython
from libc.math cimport sqrt
@cython.boundscheck(False)
@numba.jit @cython.wraparound(False)
def pairwise_python(X): def pairwise_cython(double[:, ::1] X):
M = X.shape[0] cdef int M = X.shape[0]
N = X.shape[1] cdef int N = X.shape[1]
cdef double tmp, d
D = np.empty((M, M), dtype=np.float) cdef double[:, ::1] D = np.empty((M, M),
dtype=np.float64)
for i in range(M): for i in range(M):
for j in range(M): for j in range(M):
d = 0.0 d = 0.0
for k in range(N): for k in range(N):
tmp = X[i, k] - X[j, k] tmp = X[i, k] - X[j, k]
d += tmp * tmp d += tmp * tmp
D[i, j] = np.sqrt(d) D[i, j] = sqrt(d)
return D return np.asarray(D)
The algorithmic body of each function (the nested for loops) is identical.
However the Cython code is more verbose with annotations,
both at the function definition (which we would expect for any AOT compiler),
but also within the body of the function for various utility variables.
The Numba code is just straight Python + Numpy code.
We could remove the @numba.jit decorator and step through our function with normal Python.
Example: Numpy Operations
Additionally Numba lets us use Numpy syntax directly in the function, so for example the following function is well accelerated by Numba, even though it already fits NumPy’s syntax well.
# from https://flothesof.github.io/optimizing-python-code-numpy-cython-pythran-numba.html
@numba.jit
def laplace_numba(image):
"""Laplace operator in NumPy for 2D images. Accelerated using numba."""
laplacian = ( image[:-2, 1:-1] + image[2:, 1:-1]
+ image[1:-1, :-2] + image[1:-1, 2:]
- 4*image[1:-1, 1:-1])
thresh = np.abs(laplacian) > 0.05
return thresh
Mixing and matching Numpy-style with for-loop style is often helpful when writing complex numeric algorithms.
Benchmarks in the these blogposts show that Numba is both simpler to use and often as-fast-or-faster than more commonly used technologies like Cython.
Numba drawbacks
So, given these advantages why didn’t Jake’s original prophecy hold true?
I believe that there are three primary reasons why Numba has not been more widely adopted among other open source projects:
- LLVM Dependency: Numba depends on LLVM, which was historically difficult to install without a system package manager (like apt-get, brew) or conda.
Library authors are not willing to exclude users that use other packaging toolchains, particularly Python’s standard tool,
pip. - Community Trust: Numba is largely developed within a single for-profit company (Anaconda Inc.) and its developers are not well known by other library maintainers.
- Lack of Interpretability: Numba’s output, LLVM, is less well understood by the community than Cython’s output, C (discussed in original-author comments in the last section)
All three of these are excellent reasons to avoid adding a dependency. Technical excellence alone is insufficient, and must be considered alongside community and long-term maintenance concerns.
- http://jakevdp.github.io/blog/2015/02/24/optimizing-python-with-numpy-and-numba/
- how difficult will it be for your users to install, read, modify, and contribute to your code? In the long run, this may be much more important than shaving a few milliseconds off the execution time
- http://stephanhoyer.com/2015/04/09/numba-vs-cython-how-to-choose/
- Cython is easier to distribute than Numba, which makes it a better option for user facing libraries
- The main issue is that it can be difficult to install Numba unless you use Conda, which is great tool, but not one everyone wants to use
- Cython is also a more stable and mature platform, whereas the features and performance of Numba are still evolving
- https://dionhaefner.github.io/2016/11/suck-less-scientific-python-part-2-efficient-number-crunching/
- Numba still only supports a subset of the Python and NumPy capabilities - and speedups are not always that dramatic.
But Numba has evolved recently
LLVM
Numba now depends on the easier-to-install library, llvmlite
which, as of a few months ago is pip installable with binary wheels on Windows, Mac, and Linux.
The llvmlite package is still a heavy-ish runtime dependency (42MB),
but that’s significantly less than large Cython libraries like Pandas or SciPy.
If your concern was about the average user’s inability to install Numba, then I think that this concern has been resolved.
Community
Numba has three community problems:
- Development of Numba has traditionally happened within the closed walls of Anaconda Inc (formerly Continuum Analytics)
- The Numba maintainers are not well known within the broader Python community
- There used to be a proprietary version, Numba Pro
This combination strongly attached Numba’s image to Continuum’s for-profit ventures, making community-oriented software maintainers understandably wary of dependence, for fear that dependence on this library might be used for Continuum’s financial gain at the expense of community users.
Things have changed significantly.
Numba Pro was abolished years ago. The funding for the project today comes more often from Anaconda Inc. consulting revenue, hardware vendors looking to ensure that Python runs as efficiently as possible on their systems, and from generous donations from the Gordon and Betty Moore foundation to ensure that Numba serves the open source Python community.
Developers outside of Anaconda Inc. now have core commit access, which forces communication to happen in public channels, notably GitHub (which was standard before) and Gitter chat (which is relatively new).
The maintainers are still fairly relatively unknown within the broader community. This isn’t due to any sort of conspiracy, but is instead due more to shyness or having interests outside of OSS. Antoine, Siu, Stan, and Stuart are all considerate, funny, and clever fellows with strong enthusiasm for compilers, OSS, and performance. They are quite responsive on the Numba mailing list should you have any questions or concerns.
If your concern was about Numba trapping users into a for-profit mode, then that seems to have been resolved years ago.
If your concern is more about not knowing who is behind the project then I encourage you to reach out. I would be surprised if you don’t walk away pleased.
The Continued Cases Against Numba
For completeness, let’s list a number of reasons why it is still quite reasonable to avoid Numba today:
- It isn’t a community standard
- Numba hasn’t attracted a wide developer base (compilers are hard), and so is probably still dependent on financial support for paid developers
- You want to speed up non-numeric code that includes classes, dicts, lists, etc. for which I need Cython or PyPy
- You want to build a library that is useful outside of Python, and so plan to build most numeric algorithms on C/C++/Fortran
- You prefer ahead-of-time compilation and want to avoid JIT times
- While
llvmliteis cheaper than LLVM, it’s still 50MB - Understanding the compiled results is hard, and you don’t have good familiarity with LLVM
Numba features we didn’t talk about
- Multi-core parallelism
- GPUs
- Run-time Specialization to the CPU you’re running on
- Easy to swap out for other JIT compilers, like PyPy, if they arise in the future
Update from the original blogpost authors
After writing the above I reached out both to Stan and Siu from Numba and to the original authors of the referenced blogposts to get some of their impressions now having the benefit of additional experience.
Here are a few choice responses:
-
Stan:
I think one of the biggest arguments against Numba still is time. Due to a massive rewrite of the code base, Numba, in its present form, is ~3 years old, which isn’t that old for a project like this. I think it took PyPy at least 5-7 years to reach a point where it was stable enough to really trust. Cython is 10 years old. People have good reason to be conservative with taking on new core dependencies.
-
Jake:
One thing I think would be worth fleshing-out a bit (you mention it in the final bullet list) is the fact that numba is kind of a black box from the perspective of the developer. My experience is that it works well for straightforward applications, but when it doesn’t work well it’s *extremely difficult to diagnose what the problem might be.*
Contrast that with Cython, where the html annotation output does wonders for understanding your bottlenecks both at a non-technical level (“this is dark yellow so I should do something different”) and a technical level (“let me look at the C code that was generated”). If there’s a similar tool for numba, I haven’t seen it.
-
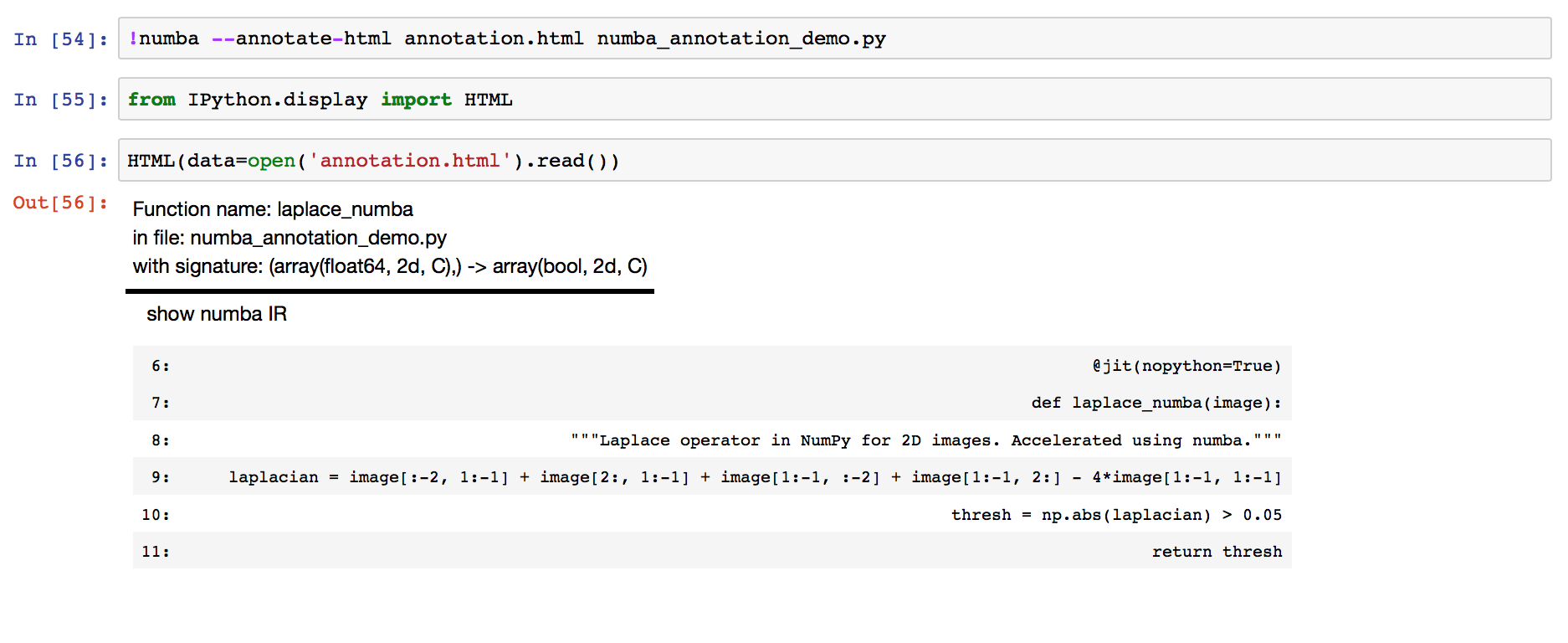
Florian:
Elaborating on Jake’s answer, I completely agree that Cython’s annotation tool does wonders in terms of understanding your code. In fact, numba does possess this too, but as a command-line utility. I tried to demonstrate this in my blogpost, but exporting the CSS in the final HTML render kind of mangles my blog post so here’s a screenshot:
This is a case where
jit(nopython=True)works, so there seems to be no coloring at all.Florian also pointed to the SciPy 2017 tutorial by Gil Forsyth and Lorena Barba
-
Dion:
I hold Numba in high regard, and the speedups impress me every time. I use it quite often to optimize some bottlenecks in our production code or data analysis pipelines (unfortunately not open source). And I love how Numba makes some functions like
scipy.optimize.minimizeorscipy.ndimage.generic_filterwell-usable with minimal effort.However, I would never use Numba to build larger systems, precisely for the reason Jake mentioned. Subjectively, Numba feels hard to debug, has cryptic error messages, and seemingly inconsistent behavior. It is not a “decorate and forget” solution; instead it always involves plenty of fiddling to get right.
That being said, if I were to build some high-level scientific library à la Astropy with some few performance bottlenecks, I would definitely favor Numba over Cython (and if it’s just to spare myself the headache of getting a working C compiler on Windows).
-
Stephan:
I wonder if there are any examples of complex codebases (say >1000 LOC) using Numba. My sense is that this is where Numba’s limitations will start to become more evident, but maybe newer features like jitclass would make this feasible.
SciPy tutorial link
As a final take-away, you might want to follow Florian’s advice and watch Gil and Lorena’s tutorial here:
blog comments powered by Disqus