Dask on HPC - Initial Work
This work is supported by Anaconda Inc. and the NSF EarthCube program.
We recently announced a collaboration between the National Center for Atmospheric Research (NCAR), Columbia University, and Anaconda Inc to accelerate the analysis of atmospheric and oceanographic data on high performance computers (HPC) with XArray and Dask. The full text of the proposed work is available here. We are very grateful to the NSF EarthCube program for funding this work, which feels particularly relevant today in the wake (and continued threat) of the major storms Harvey, Irma, and Jose.
This is a collaboration of academic scientists (Columbia), infrastructure stewards (NCAR), and software developers (Anaconda and Columbia and NCAR) to scale current workflows with XArray and Jupyter onto big-iron HPC systems and peta-scale datasets. In the first week after the grant closed a few of us focused on the quickest path to get science groups up and running with XArray, Dask, and Jupyter on these HPC systems. This blogpost details what we achieved and some of the new challenges that we’ve found in that first week. We hope to follow this blogpost with many more to come in the future. Today we cover the following topics:
- Deploying Dask with MPI
- Interactive deployments on a batch job scheduler, in this case PBS
- The virtues of JupyterLab in a remote system
- Network performance and 3GB/s infiniband
- Modernizing XArray’s interactions with Dask’s distributed scheduler
A video walkthrough deploying Dask on XArray on an HPC system is available on YouTube and instructions for atmospheric scientists with access to the Cheyenne Supercomputer is available here.
Now lets start with technical issues:
Deploying Dask with MPI
HPC systems use job schedulers like SGE, SLURM, PBS, LSF, and others. Dask has been deployed on all of these systems before either by academic groups or financial companies. However every time we do this it’s a little different and generally tailored to a particular cluster.
We wanted to make something more general. This started out as a GitHub issue on PBS scripts that tried to make a simple common template that people could copy-and-modify. Unfortunately, there were significant challenges with this. HPC systems and their job schedulers seem to focus and easily support only two common use cases:
- Embarrassingly parallel “run this script 1000 times” jobs. This is too simple for what we have to do.
- MPI jobs. This seemed like overkill, but is the approach that we ended up taking.
Deploying dask is somewhere between these two. It falls into the master-slave
pattern (or perhaps more appropriately coordinator-workers). We ended up
building an MPI4Py program that
launches Dask. MPI is well supported, and more importantly consistently
supported, by all HPC job schedulers so depending on MPI provides a level of
stability across machines. Now dask.distributed ships with a new dask-mpi
executable:
mpirun --np 4 dask-mpi
To be clear, Dask isn’t using MPI for inter-process communication. It’s still
using TCP. We’re just using MPI to launch a scheduler and several workers and
hook them all together. In pseudocode the dask-mpi executable looks
something like this:
from mpi4py import MPI
comm = MPI.COMM_WORLD
rank = comm.Get_rank()
if rank == 0:
start_dask_scheduler()
else:
start_dask_worker()
Socially this is useful because every cluster management team knows how to support MPI, so anyone with access to such a cluster has someone they can ask for help. We’ve successfully translated the question “How do I start Dask?” to the question “How do I run this MPI program?” which is a question that the technical staff at supercomputer facilities are generally much better equipped to handle.
Working Interactively on a Batch Scheduler
Our collaboration is focused on interactive analysis of big datasets. This means that people expect to open up Jupyter notebooks, connect to clusters of many machines, and compute on those machines while they sit at their computer.
Unfortunately most job schedulers were designed for batch scheduling. They will try to run your job quickly, but don’t mind waiting for a few hours for a nice set of machines on the super computer to open up. As you ask for more time and more machines, waiting times can increase drastically. For most MPI jobs this is fine because people aren’t expecting to get a result right away and they’re certainly not interacting with the program, but in our case we really do want some results right away, even if they’re only part of what we asked for.
Handling this problem long term will require both technical work and policy decisions. In the short term we take advantage of two facts:
- Many small jobs can start more quickly than a few large ones. These take advantage of holes in the schedule that are too small to be used by larger jobs.
- Dask doesn’t need to be started all at once. Workers can come and go.
And so I find that if I ask for several single machine jobs I can easily cobble together a sizable cluster that starts very quickly. In practice this looks like the following:
$ qsub start-dask.sh # only ask for one machine
$ qsub add-one-worker.sh # ask for one more machine
$ qsub add-one-worker.sh # ask for one more machine
$ qsub add-one-worker.sh # ask for one more machine
$ qsub add-one-worker.sh # ask for one more machine
$ qsub add-one-worker.sh # ask for one more machine
$ qsub add-one-worker.sh # ask for one more machine
Our main job has a wall time of about an hour. The workers have shorter wall times. They can come and go as needed throughout the computation as our computational needs change.
Jupyter Lab and Web Frontends
Our scientific collaborators enjoy building Jupyter notebooks of their work. This allows them to manage their code, scientific thoughts, and visual outputs all at once and for them serves as an artifact that they can share with their scientific teams and collaborators. To help them with this we start a Jupyter server on the same machine in their allocation that is running the Dask scheduler. We then provide them with SSH-tunneling lines that they can copy-and-paste to get access to the Jupyter server from their personal computer.
We’ve been using the new Jupyter Lab rather than the classic notebook. This is especially convenient for us because it provides much of the interactive experience that they lost by not working on their local machine. They get a file browser, terminals, easy visualization of textfiles and so on without having to repeatedly SSH into the HPC system. We get all of this functionality on a single connection and with an intuitive Jupyter interface.
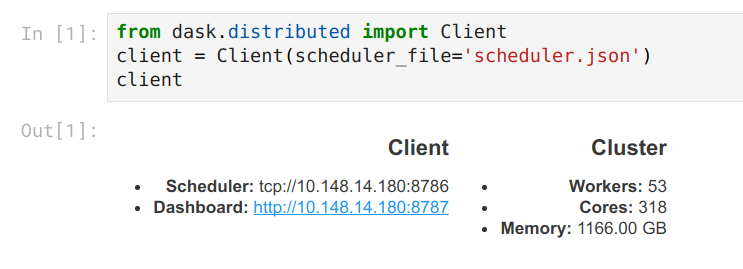
For now we give them a script to set all of this up. It starts Jupyter Lab using Dask and then prints out the SSH-tunneling line.
from dask.distributed import Client
client = Client(scheduler_file='scheduler.json')
import socket
host = client.run_on_scheduler(socket.gethostname)
def start_jlab(dask_scheduler):
import subprocess
proc = subprocess.Popen(['jupyter', 'lab', '--ip', host, '--no-browser'])
dask_scheduler.jlab_proc = proc
client.run_on_scheduler(start_jlab)
print("ssh -N -L 8787:%s:8787 -L 8888:%s:8888 -L 8789:%s:8789 cheyenne.ucar.edu" % (host, host, host))
Long term we would like to switch to an entirely point-and-click interface (perhaps something like JupyterHub) but this will requires additional thinking about deploying distributed resources along with the Jupyter server instance.
Network Performance on Infiniband
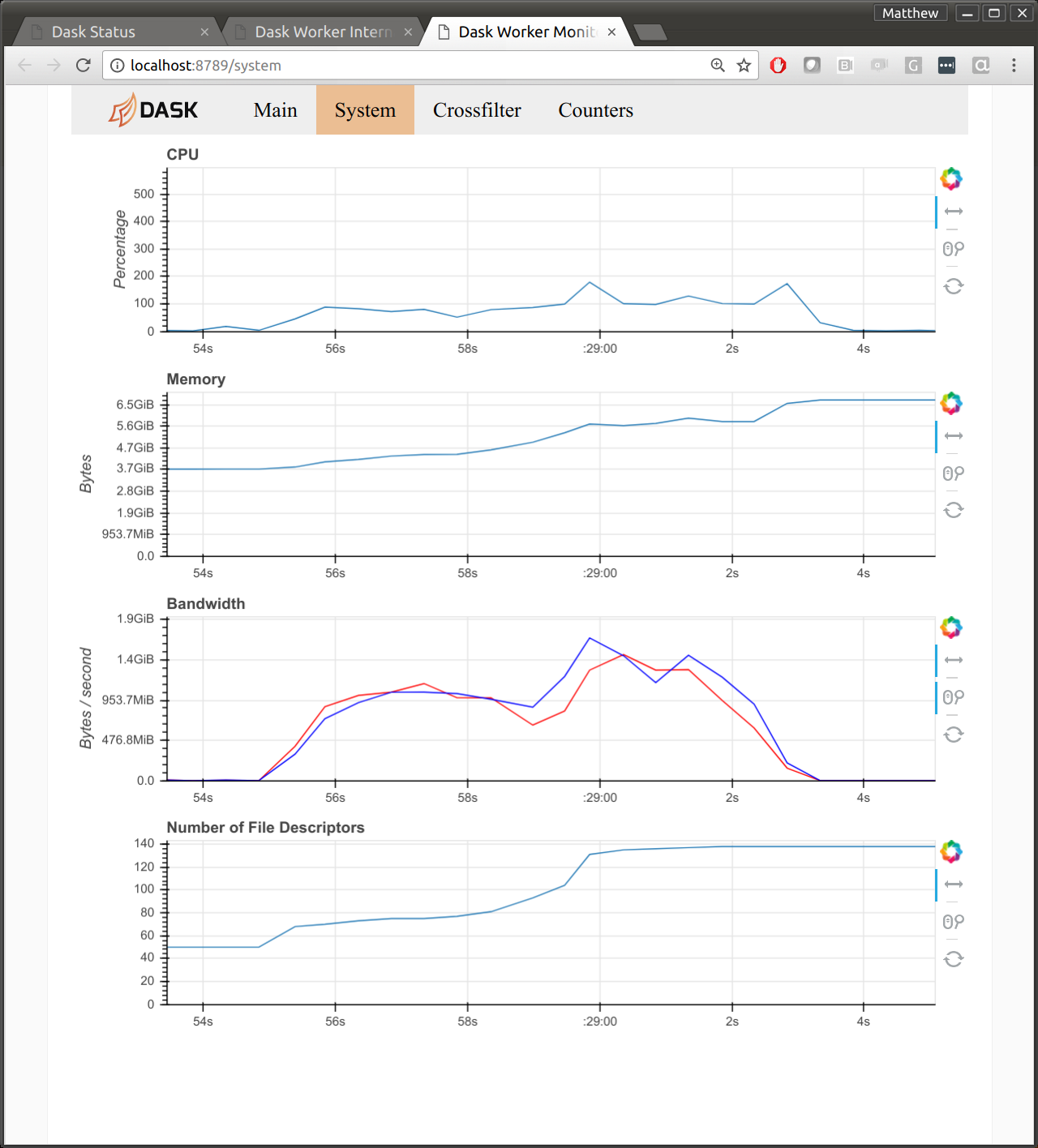
The intended computations move several terabytes across the cluster. On this cluster Dask gets about 1GB/s simultaneous read/write network bandwidth per machine using the high-speed Infiniband network. For any commodity or cloud-based system this is very fast (about 10x faster than what I observe on Amazon). However for a super-computer this is only about 30% of what’s possible (see hardware specs).
I suspect that this is due to byte-handling in Tornado, the networking library that Dask uses under the hood. The following image shows the diagnostic dashboard for one worker after a communication-heavy workload. We see 1GB/s for both read and write. We also see 100% CPU usage.
Network performance is a big question for HPC users looking at Dask. If we can get near MPI bandwidth then that may help to reduce concerns for this performance-oriented community.
How do I use Infiniband network with Dask?
XArray and Dask.distributed
XArray was the first major project to use Dask internally. This early integration was critical to prove out Dask’s internals with user feedback. However it also means that some parts of XArray were designed well before some of the newer parts of Dask, notably the asynchronous distributed scheduling features.
XArray can still use Dask on a distributed cluster, but only with the subset of features that are also available with the single machine scheduler. This means that persisting data in distributed RAM, parallel debugging, publishing shared datasets, and so on all require significantly more work today with XArray than they should.
To address this we plan to update XArray to follow a newly proposed Dask interface. This is complex enough to handle all Dask scheduling features, but light weight enough not to actually require any dependence on the Dask library itself. (Work by Jim Crist.)
We will also eventually need to look at reducing overhead for inspecting several NetCDF files, but we haven’t yet run into this, so I plan to wait.
Future Work
We think we’re at a decent point for scientific users to start playing with the system. We have a Getting Started with Dask on Cheyenne wiki page that our first set of guinea pig users have successfully run through without much trouble. We’ve also identified a number of issues that the software developers can work on while the scientific teams spin up.
- Zero copy Tornado writes to improve network bandwidth
- Enable Dask.distributed features in XArray by formalizing dask’s expected interface
- Dynamic deployments on batch job schedulers
We would love to engage other collaborators throughout this process. If you or your group work on related problems we would love to hear from you. This grant isn’t just about serving the scientific needs of researchers at Columbia and NCAR, but about building long-term systems that can benefit the entire atmospheric and oceanographic community. Please engage on the Pangeo GitHub issue tracker.
blog comments powered by Disqus