Dask Release 0.13.0
This work is supported by Continuum Analytics the XDATA Program and the Data Driven Discovery Initiative from the Moore Foundation
Summary
Dask just grew to version 0.13.0. This is a signifcant release for arrays, dataframes, and the distributed scheduler. This blogpost outlines some of the major changes since the last release November 4th.
- Python 3.6 support
- Algorithmic and API improvements for DataFrames
- Dataframe to Array conversions for Machine Learning
- Parquet support
- Scheduling Performance and Worker Rewrite
- Pervasive Visual Diagnostics with Embedded Bokeh Servers
- Windows continuous integration
- Custom serialization
You can install new versions using Conda or Pip
conda install -c conda-forge dask distributed
or
pip install dask[complete] distributed --upgrade
Python 3.6 Support
Dask and all necessary dependencies are now available on Conda Forge for Python 3.6.
Algorithmic and API Improvements for DataFrames
Thousand-core Dask deployments have become significantly more common in the last few months. This has highlighted scaling issues in some of the Dask.array and Dask.dataframe algorithms, which were originally designed for single workstations. Algorithmic and API changes can be grouped into the following two categories:
- Filling out the Pandas API
- Algorithms that needed to be changed or added due to scaling issues
Dask Dataframes now include a fuller set of the Pandas API, including the following:
- Inplace operations like
df['x'] = df.y + df.z - The full Groupby-aggregate syntax like
df.groupby(...).aggregate({'x': 'sum', 'y': ['min', max']}) - Resample on dataframes as well as series
- Pandas’ new rolling syntax
df.x.rolling(10).mean() - And much more
Additionally, collaboration with some of the larger Dask deployments has highlighted scaling issues in some algorithms, resulting in the following improvements:
- Tree reductions for groupbys, aggregations, etc.
- Multi-output-partition aggregations for groupby-aggregations with millions of groups, drop_duplicates, etc..
- Approximate algorithms for nunique
- etc..
These same collaborations have also yielded better handling of open file descriptors, changes upstream to Tornado, and upstream changes to the conda-forge CPython recipe itself to increase the default file descriptor limit on Windows up from 512.
Dataframe to Array Conversions
You can now convert Dask dataframes into Dask arrays. This is mostly to support efforts of groups building statistics and machine learning applications, where this conversion is common. For example you can load a terabyte of CSV or Parquet data, do some basic filtering and manipulation, and then convert to a Dask array to do more numeric work like SVDs, regressions, etc..
import dask.dataframe as dd
import dask.array as da
df = dd.read_csv('s3://...') # Read raw data
x = df.values # Convert to dask.array
u, s, v = da.linalg.svd(x) # Perform serious numerics
This should help machine learning and statistics developers generally, as many of the more sophisticated algorithms can be more easily implemented with the Dask array model than can be done with distributed dataframes. This change was done specifically to support the nascent third-party dask-glm project by Chris White at Capital One.
Previously this was hard because Dask.array wanted to know the size of every chunk of data, which Dask dataframes can’t provide (because, for example, it is impossible to lazily tell how many rows are in a CSV file without actually looking through it). Now that Dask.arrays have relaxed this requirement they can also support other unknown shape operations, like indexing an array with another array.
y = x[x > 0]
Parquet Support
Dask.dataframe now supports Parquet, a columnar binary store for tabular data commonly used in distributed clusters and the Hadoop ecosystem.
import dask.dataframe as dd
df = dd.read_parquet('myfile.parquet') # Read from Parquet
df.to_parquet('myfile.parquet', compression='snappy') # Write to Parquet
This is done through the new fastparquet library, a Numba-accelerated version of the Pure Python parquet-python. Fastparquet was built and is maintained by Martin Durant. It’s also exciting to see the Parquet-cpp project gain Python support through Arrow and work by Wes McKinney and Uwe Korn. Parquet has gone from inaccessible in Python to having multiple competing implementations, which is a wonderful and exciting change for the “Big Data” Python ecosystem.
Scheduling Performance and Worker Rewrite
The internals of the distributed scheduler and workers are significantly modified. Users shouldn’t experience much change here except for general performance enhancement, more upcoming features, and much deeper visual diagnostics through Bokeh servers.
We’ve pushed some of the scheduling logic from the scheduler onto the workers. This lets us do two things:
- We keep a much larger backlog of tasks on the workers. This allows workers to optimize and saturate their hardware more effectively. As a result, complex computations end up being significantly faster.
- We can more easily deliver on a rising number of requests for complex scheduling features. For example, GPU users will be happy to learn that you can now specify abstract resource constraints like “this task requires a GPU” and “this worker has four GPUs” and the scheduler and workers will allocate tasks accordingly. This is just one example of a feature that was easy to implement after the scheduler/worker redesign and is now available.
Pervasive Visual Diagnostics with Embedded Bokeh Servers
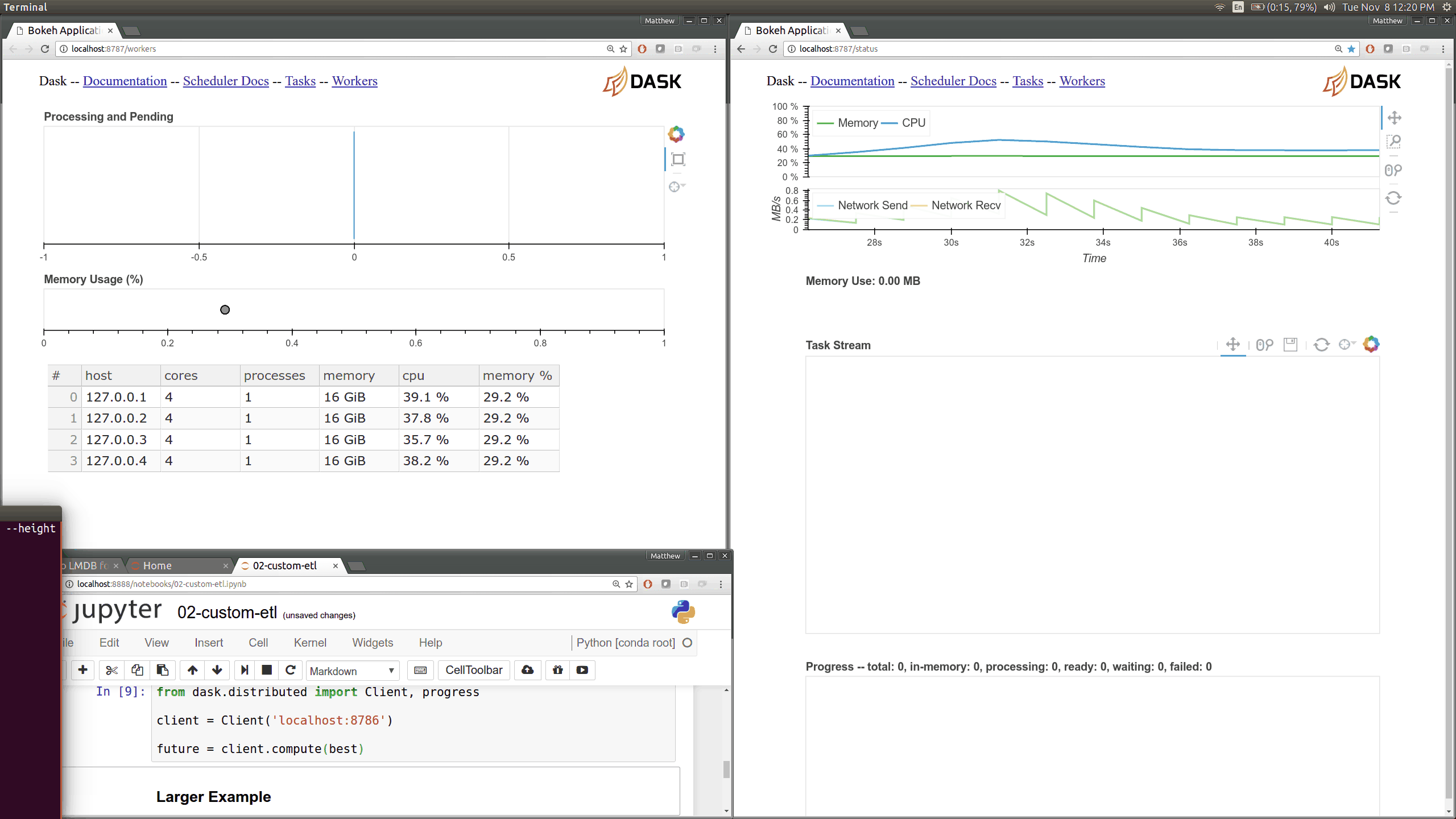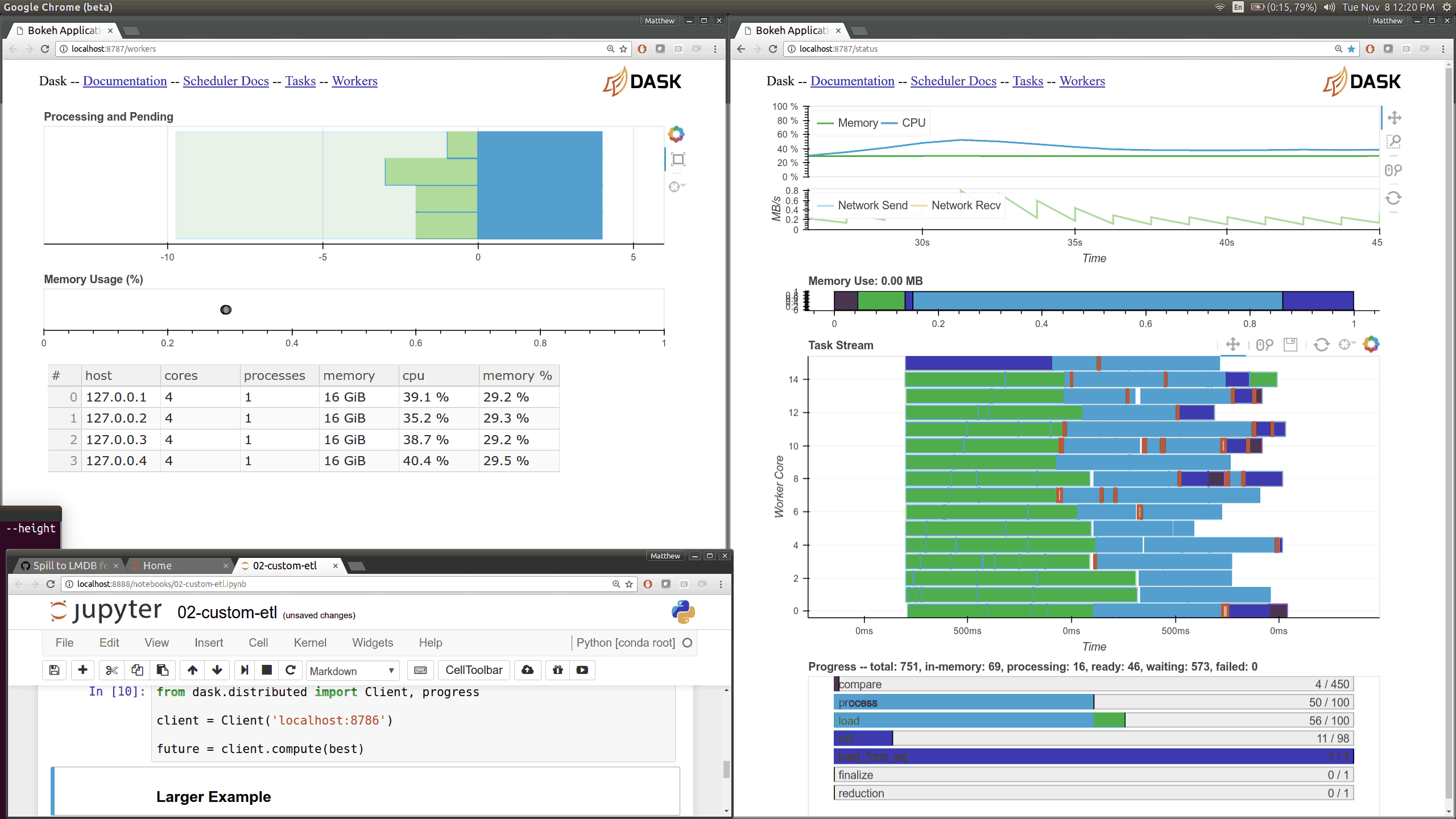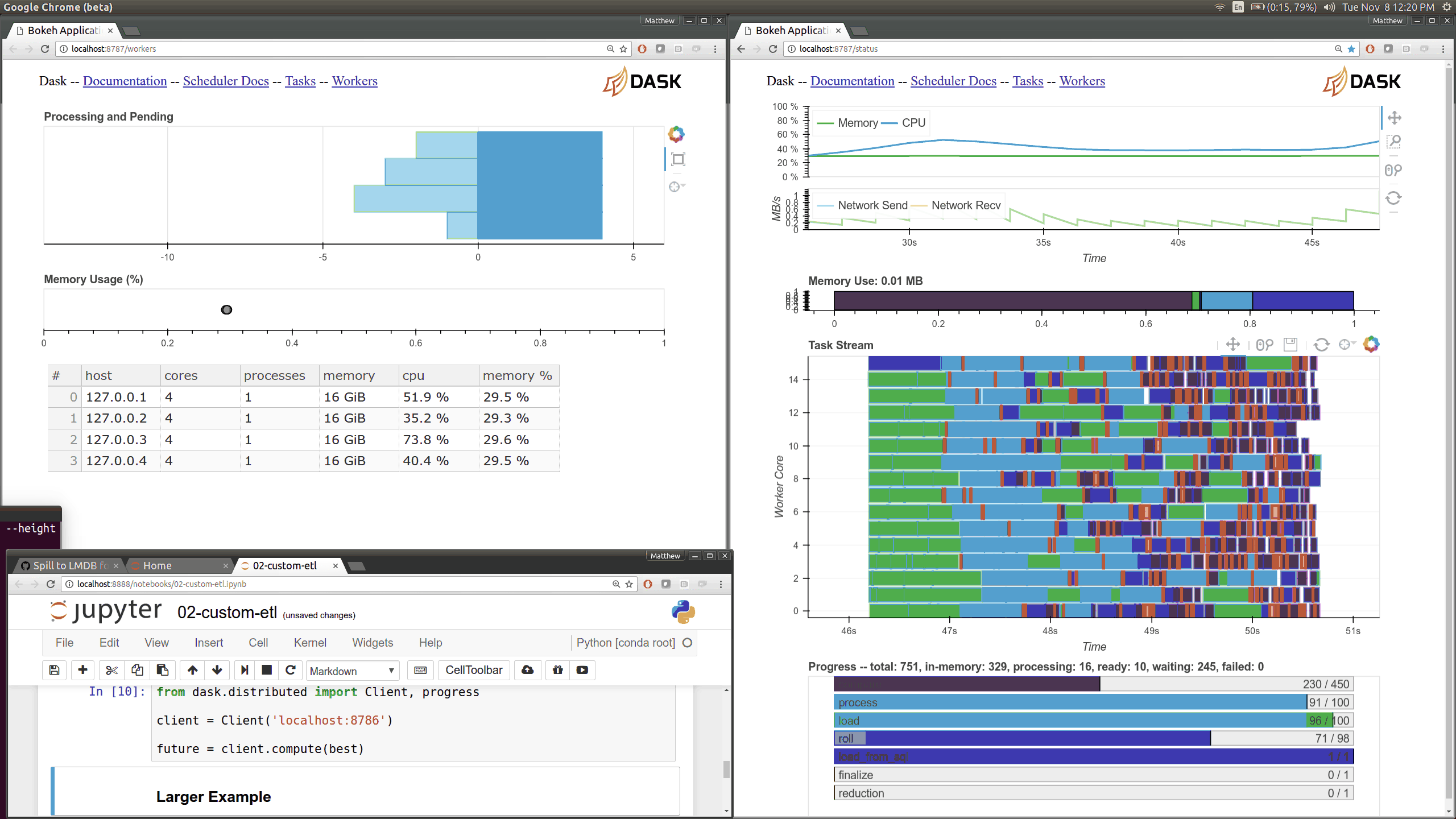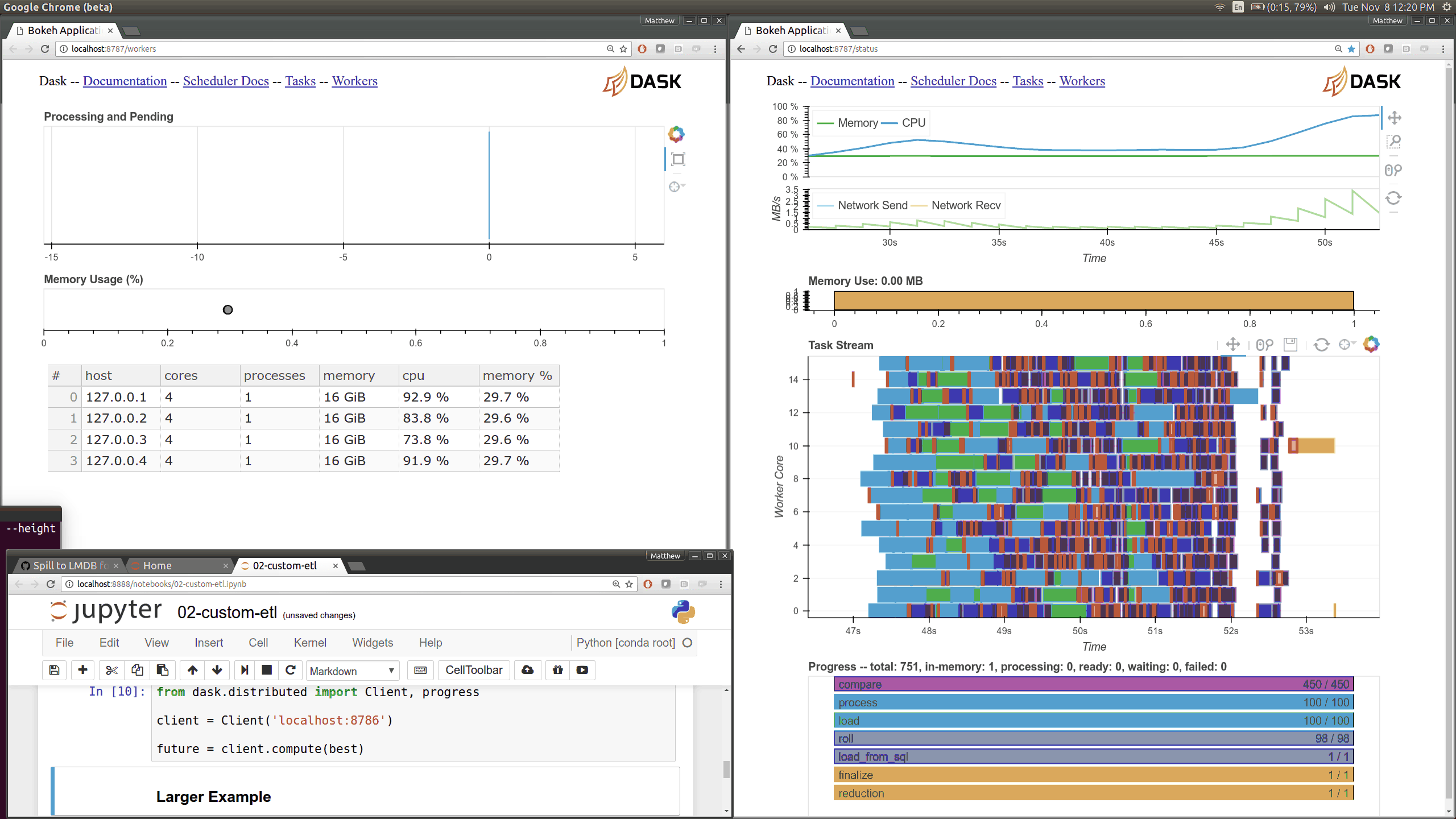
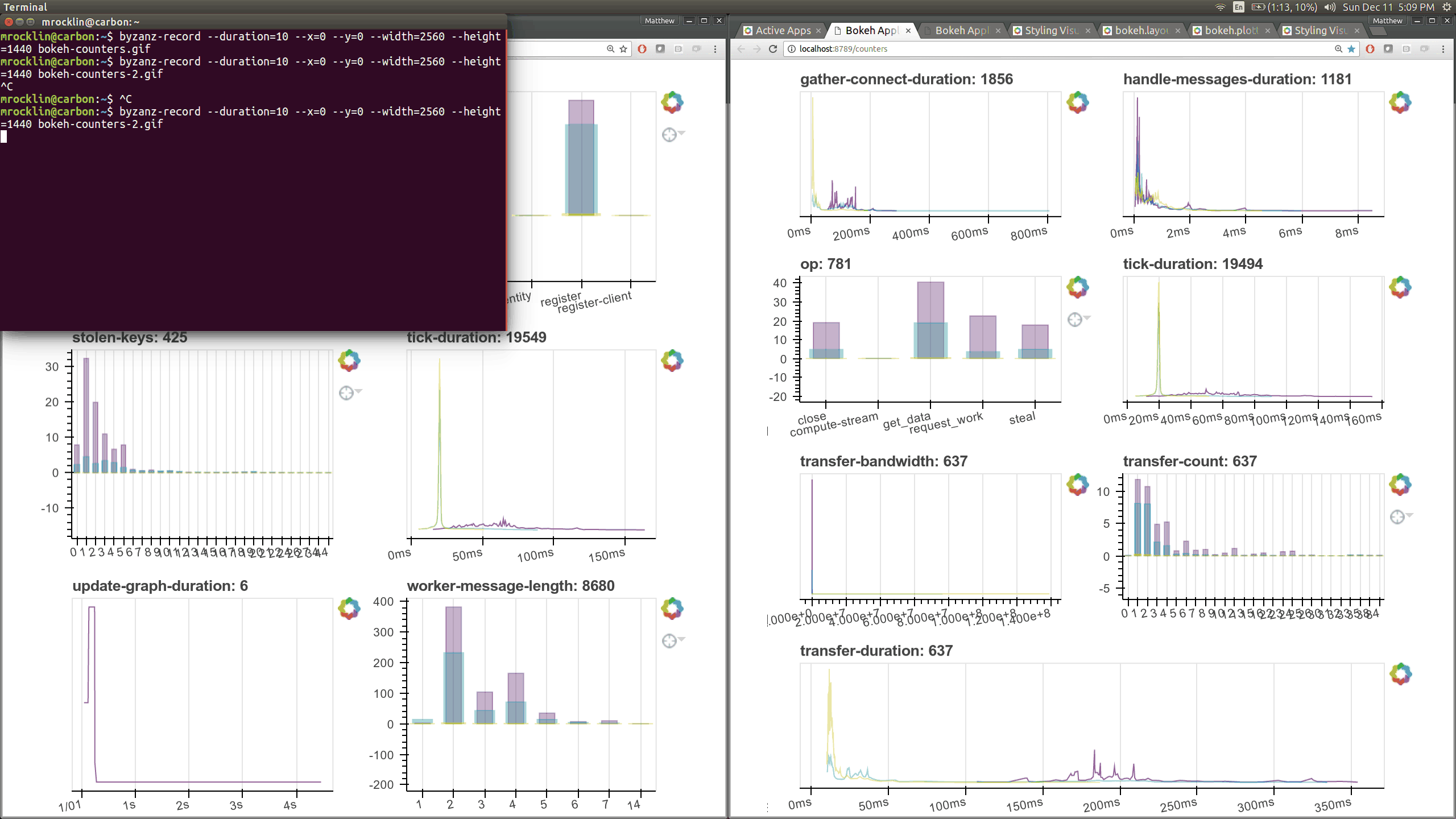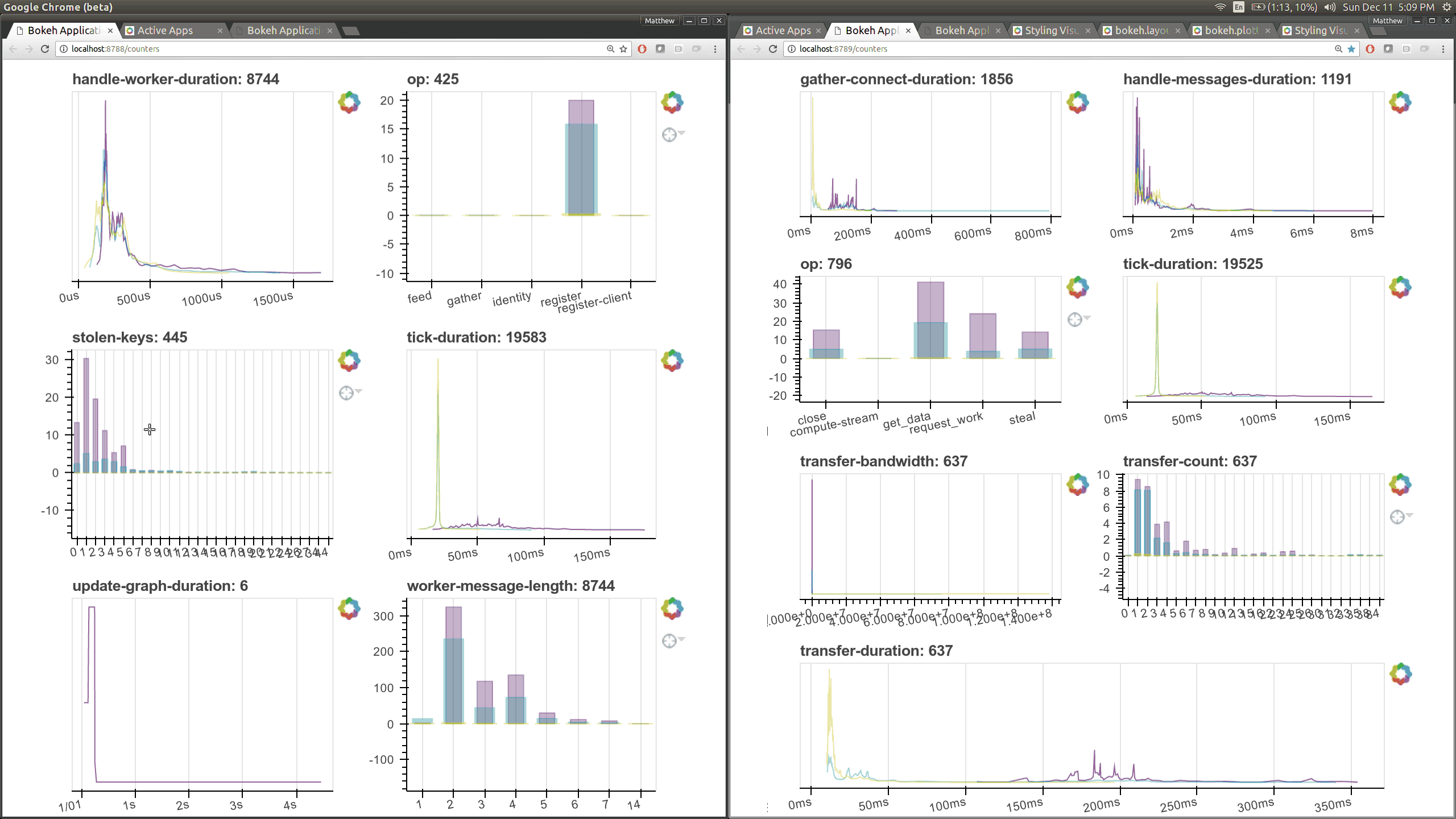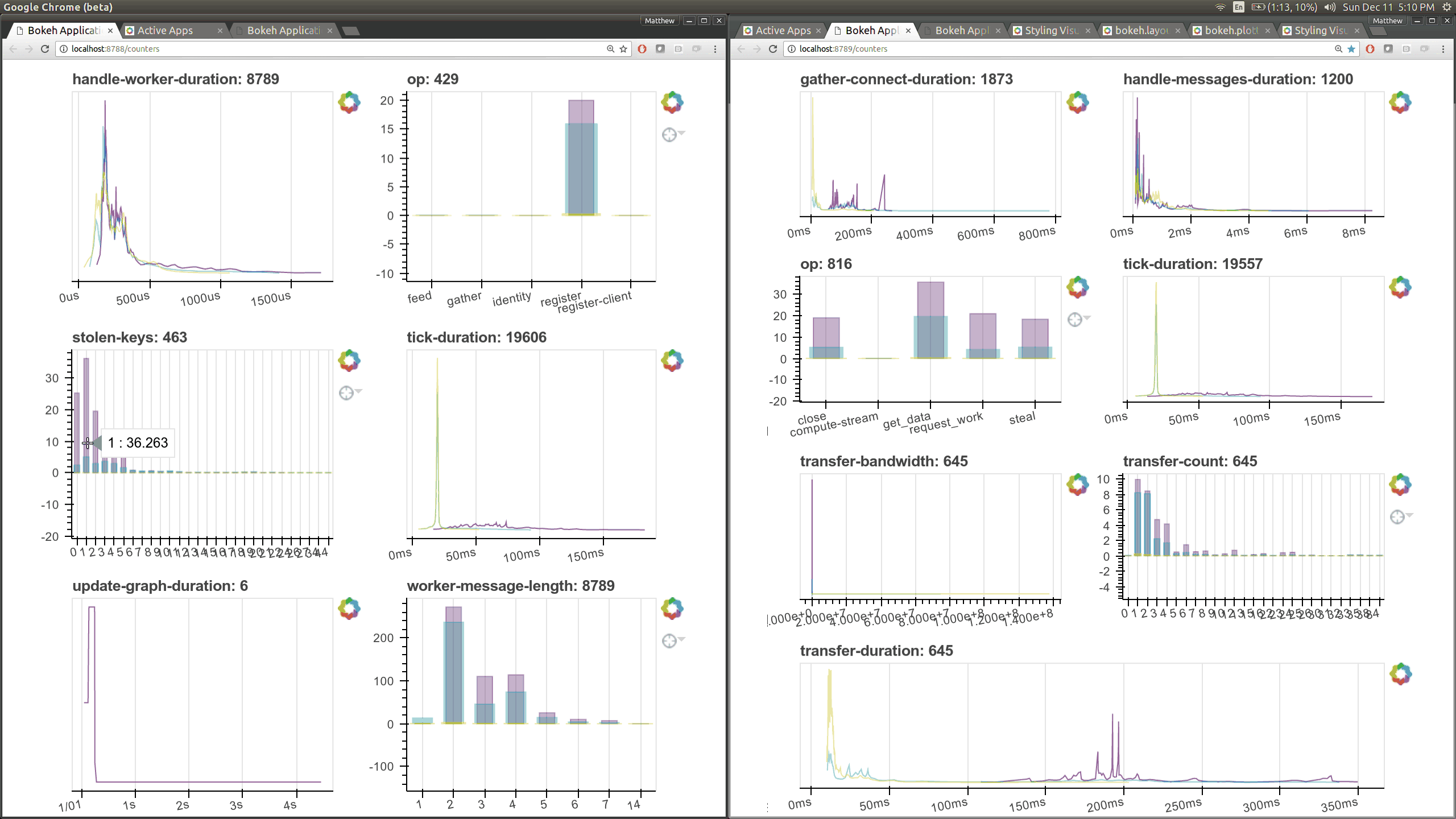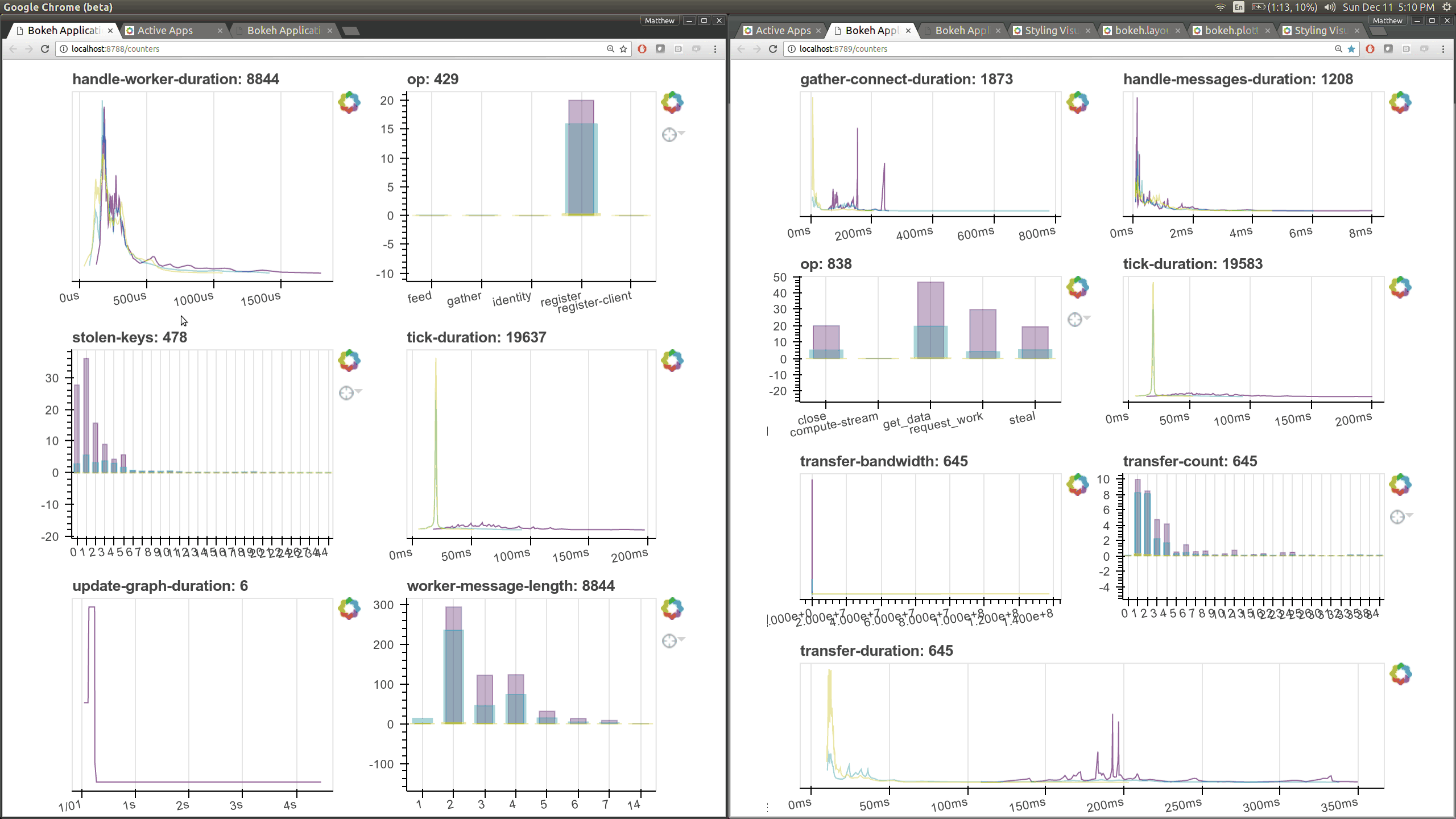
While optimizing scheduler performance we built several new visual diagnostics using Bokeh. There is now a Bokeh Server running within the scheduler and within every worker.
Current Dask.distributed users will be familiar with the current diagnostic dashboards:
These plots provide intuition about the state of the cluster and the computations currently in flight. These dashboards are generally well loved.
There are now many more of these, though more focused on internal state and timings that will be of interest to developers and power users than to a typical users. Here are a couple of the new pages (of which there are seven) that show various timings and counters of various parts of the worker and scheduler internals.
The previous Bokeh dashboards were served from a separate process that queried the scheduler periodically (every 100ms). Now there are new Bokeh servers within every worker and a new Bokeh server within the scheduler process itself rather than in a separate process. Because these servers are embedded they have direct access to the state of the scheduler and workers which significantly reduces barriers for us to build out new visuals. However, this also adds some load to the scheduler, which can often be compute bound. These pages are available at new ports, 8788 for the scheduler and 8789 for the worker by default.
Custom Serialization
This is actually a change that occurred in the last release, but I haven’t written about it and it’s important, so I’m including it here.
Previously inter-worker communication of data was accomplished with Pickle/Cloudpickle and optional generic compression like LZ4/Snappy. This was robust and worked mostly fine, but left out some exotic data types and did not provide optimal performance.
Now we can serialize different types with special consideration. This allows special types, like NumPy arrays, to pass through without unnecessary memory copies and also allows us to use more exotic data-type specific compression techniques like Blosc.
It also allows Dask to serialize some previously unserializable types. In particular this was intended to solve the Dask.array climate science community’s concern about HDF5 and NetCDF files which (correctly) are unpicklable and so restricted to single-machine use.
This is also the first step towards two frequently requested features (neither of these exist yet):
- Better support for GPU-GPU specific serialization options. We are now a large step closer to generalizing away our assumption of TCP Sockets as the universal communication mechanism.
- Passing data between workers of different runtime languages. By embracing other protocols than Pickle we begin to allow for the communication of data between workers of different software environments.
What’s Next
So what should we expect to see in the future for Dask?
- Communication: Now that workers are more fully saturated we’ve found that communication issues are arising more frequently as bottlenecks. This might be because everything else is nearing optimal or it might be because of the increased contention in the workers now that they are idle less often. Many of our new diagnostics are intended to measure components of the communication pipeline.
- Third Party Tools: We’re seeing a nice growth of utilities like dask-drmaa for launching clusters on DRMAA job schedulers (SGE, SLURM, LSF) and dask-glm for solvers for GLM-like machine-learning algorithms. I hope that external projects like these become the main focus of Dask development going forward as Dask penetrates new domains.
- Blogging: I’ll be launching a few fun blog posts throughout the next couple of weeks. Stay tuned.
Learn More
You can install or upgrade using Conda or Pip
conda install -c conda-forge dask distributed
or
pip install dask[complete] distributed --upgrade
You can learn more about Dask and its distributed scheduler at these websites:
Acknowledgements
Since the last main release the following developers have contributed to the core Dask repostiory (parallel algorithms, arrays, dataframes, etc..)
- Alexander C. Booth
- Antoine Pitrou
- Christopher Prohm
- Frederic Laliberte
- Jim Crist
- Martin Durant
- Matthew Rocklin
- Mike Graham
- Rolando (Max) Espinoza
- Sinhrks
- Stuart Archibald
And the following developers have contributed to the Dask/distributed repository (distributed scheduling, network communication, etc..)
- Antoine Pitrou
- jakirkham
- Jeff Reback
- Jim Crist
- Martin Durant
- Matthew Rocklin
- rbubley
- Stephan Hoyer
- strets123
- Travis E. Oliphant
blog comments powered by Disqus