PyData and the GIL
This work is supported by Continuum Analytics and the XDATA Program as part of the Blaze Project
tl;dr Many PyData projects release the GIL. Multi-core parallelism is alive and well.
Introduction
Machines grow more cores every year. My cheap laptop has four cores and a heavy workstation rivals a decent cluster without the hardware hassle. When I bring this up in conversation people often ask about the GIL and whether or not this poses a problem to the PyData ecosystem and shared-memory parallelism.
Q: Given the growth of shared-memory parallelism, should the PyData ecosystem be concerned about the GIL?
A: No, we should be very excited about this growth. We’re well poised to exploit it.
For those who aren’t familiar, the Global Interpreter Lock (GIL) is a CPython feature/bug that stops threads from manipulating Python objects in parallel. This cripples Pure Python shared-memory parallelism.
This sounds like a big deal but it doesn’t really affect the PyData stack (NumPy/Pandas/SciKits). Most PyData projects don’t spend much time in Python code. They spend 99% of their time in C/Fortran/Cython code. This code can often release the GIL. The following projects release the GIL at various stages:
- NumPy
- SciPy
- Numba (if requested) (example docs)
- SciKit Learn
- Anything that mostly uses the above projects
- if you add more in the comments then I will post them here
Our software stack has roots in scientific computing which has an amazing relationship with using all-of-the-hardware. I would like to see the development community lean in to the use of shared-memory parallelism. This feels like a large low-hanging fruit.
Quick Example with dask.array
As a quick example, we compute a large random dot product with
dask.array and look at top. Dask.array computes
large array operations by breaking arrays up in to many small NumPy arrays and
then executing those array operations in multiple threads.
In [1]: import dask.array as da
In [2]: x = da.random.random((10000, 10000), blockshape=(1000, 1000))
In [3]: float(x.dot(x.T).sum())
Out[3]: 250026827523.19141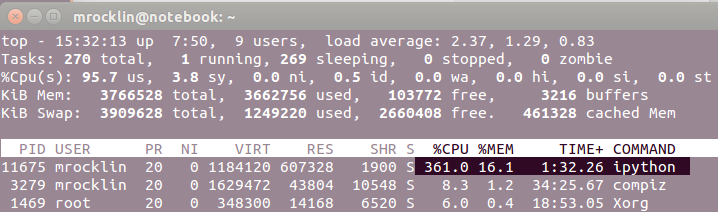
Technical note: my BLAS is set to use one thread only, the parallelism in the above example is strictly due to multiple Python worker threads, and not due to parallelism in the underlying native code.
Note the 361.0% CPU utilization in the ipython process.
Because the PyData stack is fundamentally based on native compiled code, multiple Python threads can crunch data in parallel without worrying about the GIL. The GIL does not have to affect us in a significant way.
That’s not true, the GIL hurts Python in the following cases
Text
We don’t have a good C/Fortran/Cython solution for text. When given a
pile-of-text-files we often switch from threads to processes and use the
multiprocessing module. This limits inter-worker communication but this is
rarely an issue for this kind of embarrassingly parallel work.
The multiprocessing workflow is fairly simple. I’ve written about this in the toolz docs and in a blogpost about dask.bag.
Pandas
Pandas does not yet release the GIL in computationally intensive code. It probably could though. This requires momentum from the community and some grunt-work by some of the Pandas devs. I have a small issue here and I think that Phil Cloud is looking into it.
PyData <3 Shared Memory Parallelism
If you’re looking for more speed in compute-bound applications then consider threading and heavy workstation machines. I personally find this approach to be more convenient than spinning up a cluster.
blog comments powered by Disqus