ReIntroducing Into Clean data migration (with graphs!)
This work is supported by Continuum Analytics and the XDATA Program as part of the Blaze Project
tl;dr into efficiently migrates data between formats.
Motivation
We spend a lot of time migrating data from common interchange formats, like
CSV, to efficient computation formats like an array, a database or binary
store. Worse, many don’t migrate data to efficient formats because they don’t
know how or can’t manage the particular migration process for their tools.
Your choice of data format is important. It strongly impacts performance (10x is a good rule of thumb) and who can easily use and interpret your data.
When advocating for Blaze I often say “Blaze can help you query your data in a variety of formats.” This assumes that you’re able to actually get it in to that format.
Enter the into project
The into function efficiently migrates data between formats.
These formats include both in-memory data structures like the following:
list, set, tuple, Iterator
numpy.ndarray, pandas.DataFrame, dynd.array
Streaming Sequences of any of the above
as well as persistent data living outside of Python like the following:
CSV, JSON, line-delimited-JSON
Remote versions of the above
HDF5 (both standard and Pandas formatting), BColz, SAS
SQL databases (anything supported by SQLAlchemy), Mongo
The into project migrates data between any pair of these formats efficiently
by using a network of pairwise conversions. (visualized towards the bottom of
this post)
How to use it
The into function takes two arguments, a source and a target. It moves data
in the source to the target. The source and target can take the following
forms
| Target | Source | Example |
| Object | Object | A particular DataFrame or list |
| String | String | 'file.csv', 'postgresql://hostname::tablename' |
| Type | Like list or pd.DataFrame |
So the following would be valid calls to into
>>> into(list, df) # create new list from Pandas DataFrame
>>> into([], df) # append onto existing list
>>> into('myfile.json', df) # Dump dataframe to line-delimited JSON
>>> into(Iterator, 'myfiles.*.csv') # Stream through many CSV files
>>> into('postgresql://hostname::tablename', df) # Migrate dataframe to Postgres
>>> into('postgresql://hostname::tablename', 'myfile.*.csv') # Load CSVs to Postgres
>>> into('myfile.json', 'postgresql://hostname::tablename') # Dump Postgres to JSON
>>> into(pd.DataFrame, 'mongodb://hostname/db::collection') # Dump Mongo to DataFrameNote that into is a single function. We’re used to doing this with various
to_csv, from_sql methods on various types. The into api is very small;
Here is what you need in order to get started:
$ pip install into
>>> from into import intoExamples
We now show some of those same examples in more depth.
Turn list into numpy array
>>> import numpy as np
>>> into(np.ndarray, [1, 2, 3])
array([1, 2, 3])Load CSV file into Python list
>>> into(list, 'accounts.csv')
[(1, 'Alice', 100),
(2, 'Bob', 200),
(3, 'Charlie', 300),
(4, 'Denis', 400),
(5, 'Edith', 500)]Translate CSV file into JSON
>>> into('accounts.json', 'accounts.csv')$ head accounts.json
{"balance": 100, "id": 1, "name": "Alice"}
{"balance": 200, "id": 2, "name": "Bob"}
{"balance": 300, "id": 3, "name": "Charlie"}
{"balance": 400, "id": 4, "name": "Denis"}
{"balance": 500, "id": 5, "name": "Edith"}
Translate line-delimited JSON into a Pandas DataFrame
>>> import pandas as pd
>>> into(pd.DataFrame, 'accounts.json')
balance id name
0 100 1 Alice
1 200 2 Bob
2 300 3 Charlie
3 400 4 Denis
4 500 5 EdithHow does it work?
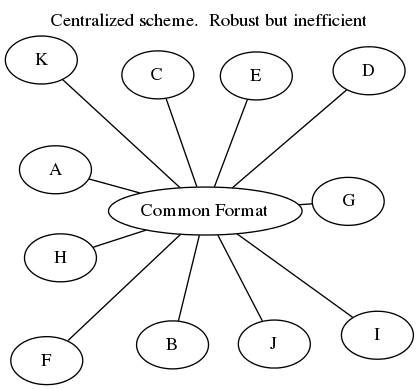
This is challenging. Robust and efficient conversions between any two pairs of
formats is fraught with special cases and bizarre libraries. The common
solution is to convert through a common format like a DataFrame, or streaming
in-memory lists, dicts, etc. (see dat)
or through a serialization format like
ProtoBuf or
Thrift. These are excellent options and often
what you want. Sometimes however this can be slow, particularly when dealing
with live computational systems or with finicky storage solutions.
Consider for example, migrating between a numpy.recarray and a
pandas.DataFrame. We can migrate this data very quickly in place. The bytes
of data don’t need to change, only the metadata surrounding them. We don’t
need to serialize to an interchange format or translate to intermediate
pure Python objects.
Consider migrating data from a CSV file to a PostgreSQL database. Using Python iterators through SQLAlchemy we rarely exceed migration speeds greater than 2000 records per second. However using direct CSV loaders native to PostgreSQL we can achieve speeds greater than 50000 records per second. This is the difference between an overnight job and a cup of coffee. However this requires that we’re flexible enough to use special code in special situations.
Expert pairwise interactions are often an order of magnitude faster than generic solutions.
Into is a network of these pairwise migrations. We visualize that network below:
Into’s decentralized migration scheme. Complex but powerful</figcaption> </figure>
Each node is a data format. Each directed edge is a function that transforms
data between two formats. A single call to into may traverse multiple edges
and multiple intermediate formats. For example, we when migrate a CSV file to
a Mongo database we might take the following route:
- Load in to a
DataFrame(pandas.read_csv) - Convert to
np.recarray(DataFrame.to_records) - Then to a Python
Iterator(np.ndarray.tolist) - Finally to Mongo (
pymongo.Collection.insert)
Alternatively we could write a special function that uses MongoDB’s native CSV
loader and shortcut this entire process with a direct edge CSV -> Mongo.
To find the most efficient route we weight the edges of this network with
relative costs (measured ad-hoc.) We use networkx to find the shortest path
before we start the migration. If for some reason an edge fails (raises
NotImplementedError) we can reroute automatically. In this way we’re both
efficient and robust to failure.
Note that we color some nodes red. These nodes can be larger than memory.
When we migrate between two red nodes (both the input and output may be larger
than memory) then we limit our path to the red subgraph to ensure that we don’t
blow up mid-migration. One format to note is chunks(...) like
chunks(DataFrame) which is an iterable of in-memory DataFrames. This
convenient meta-format allows us to use compact data structures like numpy
arrays and pandas DataFrames on large data while keeping only a few tens of
megabytes in memory at a time.
The networked approach allows developers to write specialized code for special situations and know that this code will only be used in the right situation. This approach allows us to handle a very complex problem in an isolated and separable manner. The central dispatching system keeps us sane.
History
I wrote about into long ago in connection to Blaze. I then promptly shut up about it. This was because the old implementation (before the network approach) was difficult to extend/maintain and wasn’t ready for prime-time.
I am very happy with this network. Unexpected applications very often just
work and into is now ready for prime-time. It’s also available independently
from Blaze, both via conda and via pip. The
major dependencies are NumPy, Pandas, and NetworkX so it’s relatively
lightweight for most people who read my blog. If you want to take advantage of
some of the higher performing formats, like HDF5, you’ll need to install
those libraries as well (pro-tip, use conda).
How do I get started?
You should download a recent version.
$ pip install --upgrade git+https://github.com/ContinuumIO/into
or
$ conda install into --channel blaze
You then might want to go through the first half of this tutorial
Or read the docs.
Or just give it a shot without reading anything. My hope is that the interface
is simple enough (just one function!) that users can pick it up naturally. If
you run in to issues then I’d love to hear about them at
blaze-dev@continuum.io
blog comments powered by Disqus
