Efficiently Store Pandas DataFrames
This work is supported by Continuum Analytics and the XDATA Program as part of the Blaze Project
tl;dr We benchmark several options to store Pandas DataFrames to disk. Good options exist for numeric data but text is a pain. Categorical dtypes are a good option.
Introduction
For dask.frame I need to read and write Pandas DataFrames to disk. Both disk bandwidth and serialization speed limit storage performance.
- Disk bandwidth, between 100MB/s and 800MB/s for a notebook hard drive, is limited purely by hardware. Not much we can do here except buy better drives.
- Serialization cost though varies widely by library and context. We can be smart here. Serialization is the conversion of a Python variable (e.g. DataFrame) to a stream of bytes that can be written raw to disk.
Typically we use libraries like pickle to serialize Python objects. For
dask.frame we really care about doing this quickly so we’re going to also
look at a few alternatives.
Contenders
pickle- The standard library pure Python solutioncPickle- The standard library C solutionpickle.dumps(data, protocol=2)- pickle and cPickle support multiple protocols. Protocol 2 is good for numeric data.json- using the standardlibjsonlibrary, we encode the values and index as lists of ints/stringsjson-no-index- Same as above except that we don’t encode the index of the DataFrame, e.g.0, 1, ...We’ll find that JSON does surprisingly well on pure text data.- msgpack - A binary JSON alternative
CSV- The venerablepandas.read_csvandDataFrame.to_csvhdfstore- Pandas’ custom HDF5 storage format
Additionally we mention but don’t include the following:
dillandcloudpickle- formats commonly used for function serialization. These perform about the same ascPicklehickle- A pickle interface over HDF5. This does well on NumPy data but doesn’t support Pandas DataFrames well.
Experiment
Disclaimer: We’re about to issue performance numbers on a toy dataset. You should not trust that what follows generalizes to your data. You should look at your own data and run benchmarks yourself. My benchmarks lie.
We create a DataFrame with two columns, one with numeric data, and one with text. The text column has repeated values (1000 unique values, each repeated 1000 times) while the numeric column is all unique. This is fairly typical of data that I see in the wild.
df = pd.DataFrame({'text': [str(i % 1000) for i in range(1000000)],
'numbers': range(1000000)})Now we time the various dumps and loads methods of the different
serialization libraries and plot the results below.
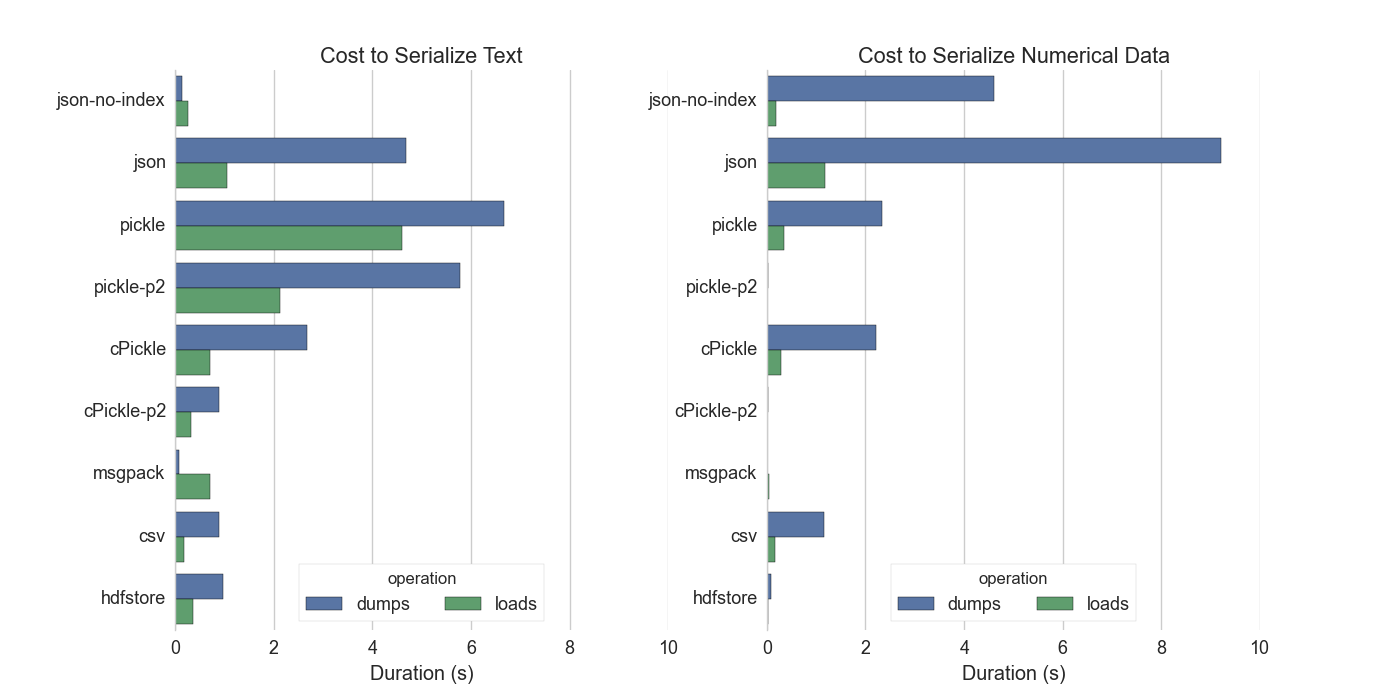
As a point of reference writing the serialized result to disk and reading it back again should take somewhere between 0.05s and 0.5s on standard hard drives. We want to keep serialization costs below this threshold.
Thank you to Michael Waskom for making those charts (see twitter conversation and his alternative charts)
Gist to recreate plots here: https://gist.github.com/mrocklin/4f6d06a2ccc03731dd5f
Further Disclaimer: These numbers average from multiple repeated calls to
loads/dumps. Actual performance in the wild is likely worse.
Observations
We have good options for numeric data but not for text. This is unfortunate; serializing ASCII text should be cheap. We lose here because we store text in a Series with the NumPy dtype ‘O’ for generic Python objects. We don’t have a dedicated variable length string dtype. This is tragic.
For numeric data the successful systems systems record a small amount of
metadata and then dump the raw bytes. The main takeaway from this is that you
should use the protocol=2 keyword argument to pickle. This option isn’t
well known but strongly impacts preformance.
Note: Aaron Meurer notes in the comments that for Python 3 users protocol=3
is already default. Python 3 users can trust the default protocol= setting
to be efficient and should not specify protocol=2.
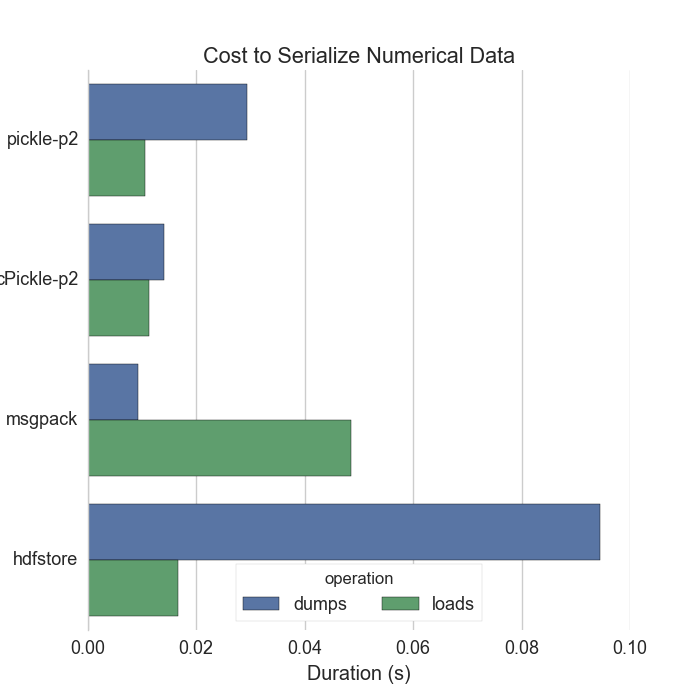
Some thoughts on text
-
Text should be easy to serialize. It’s already text!
-
JSON-no-index serializes the text values of the dataframe (not the integer index) as a list of strings. This assumes that the data are strings which is why it’s able to outperform the others, even though it’s not an optimized format. This is what we would gain if we had a
stringdtype rather than relying on the NumPy Object dtype,'O'. -
MsgPack is surpsingly fast compared to cPickle
-
MsgPack is oddly unbalanced, it can dump text data very quickly but takes a while to load it back in. Can we improve msgpack load speeds?
-
CSV text loads are fast. Hooray for
pandas.read_csv.
Some thoughts on numeric data
-
Both
pickle(..., protocol=2)andmsgpackdump raw bytes. These are well below disk I/O speeds. Hooray! -
There isn’t much reason to compare performance below this level.
Categoricals to the Rescue
Pandas recently added support for categorical
data. We
use categorical data when our values take on a fixed number of possible options
with potentially many repeats (like stock ticker symbols.) We enumerate these
possible options (AAPL: 1, GOOG: 2, MSFT: 3, ...) and use those numbers
in place of the text. This works well when there are many more
observations/rows than there are unique values. Recall that in our case we
have one million rows but only one thousand unique values. This is typical for
many kinds of data.
This is great! We’ve shrunk the amount of text data by a factor of a thousand, replacing it with cheap-to-serialize numeric data.
>>> df['text'] = df['text'].astype('category')
>>> df.text
0 0
1 1
2 2
3 3
...
999997 997
999998 998
999999 999
Name: text, Length: 1000000, dtype: category
Categories (1000, object): [0 < 1 < 10 < 100 ... 996 < 997 < 998 < 999]Lets consider the costs of doing this conversion and of serializing it afterwards relative to the costs of just serializing it.
| seconds | |
|---|---|
| Serialize Original Text | 1.042523 |
| Convert to Categories | 0.072093 |
| Serialize Categorical Data | 0.028223 |
When our data is amenable to categories then it’s cheaper to convert-then-serialize than it is to serialize the raw text. Repeated serializations are just pure-win. Categorical data is good for other reasons too; computations on object dtype in Pandas generally happen at Python speeds. If you care about performance then categoricals are definitely something to roll in to your workflow.
Final Thoughts
- Several excellent serialization options exist, each with different strengths.
- A combination of good serialization support for numeric data and Pandas categorical dtypes enable efficient serialization and storage of DataFrames.
- Object dtype is bad for PyData. String dtypes would be nice. I’d like to shout out to DyND a possible NumPy replacement that would resolve this.
- MsgPack provides surprisingly good performance over custom Python solutions, why is that?
- I suspect that we could improve performance by special casing Object dtypes and assuming that they contain only text.
blog comments powered by Disqus